| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:29:06 UTC |
|---|
| Update Date | 2020-04-21 18:12:10 UTC |
|---|
| BMDB ID | BMDB0000351 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 3-Hydroxy-2-methyl-[R-(R,S)]-butanoic acid |
|---|
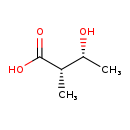
| Description | (2S,3R)-3-Hydroxy-2-methylbutanoic acid, also known as 3-hydroxy-2-methyl-[R-(r*,s*)]-butanoate or (2S,3R)-nilic acid, belongs to the class of organic compounds known as hydroxy fatty acids. These are fatty acids in which the chain bears a hydroxyl group. Based on a literature review very few articles have been published on (2S,3R)-3-Hydroxy-2-methylbutanoic acid. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S,3R)-3-Hydroxy-2-methylbutanoate | Generator | | 3-Hydroxy-2-methyl-[R-(r*,s*)]-butanoate | HMDB | | 3-Hydroxy-2-methyl-[R-(r*,s*)]-butanoic acid | HMDB | | 3-Hydroxy-2-methyl-[R-(R,S)]-butanoate | HMDB | | (2S,3R)-3-Hydroxy-2-methylbutyric acid | HMDB | | (2S,3R)-Nilic acid | HMDB | | 3-Hydroxy-2-methyl-[R-(R,S)]-butanoic acid | HMDB | | HMBA | HMDB | | (2S,3R)-3-Hydroxy-2-methylbutanoic acid | HMDB |
|
|---|
| Chemical Formula | C5H10O3 |
|---|
| Average Molecular Weight | 118.1311 |
|---|
| Monoisotopic Molecular Weight | 118.062994186 |
|---|
| IUPAC Name | (2S,3R)-3-hydroxy-2-methylbutanoic acid |
|---|
| Traditional Name | (2S,3R)-3-hydroxy-2-methylbutanoic acid |
|---|
| CAS Registry Number | 71526-30-2 |
|---|
| SMILES | C[C@@H](O)[C@H](C)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H10O3/c1-3(4(2)6)5(7)8/h3-4,6H,1-2H3,(H,7,8)/t3-,4+/m0/s1 |
|---|
| InChI Key | VEXDRERIMPLZLU-IUYQGCFVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hydroxy fatty acids. These are fatty acids in which the chain bears a hydroxyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Hydroxy fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Beta-hydroxy acid
- Branched fatty acid
- Hydroxy fatty acid
- Short-chain hydroxy acid
- Methyl-branched fatty acid
- Hydroxy acid
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organooxygen compound
- Alcohol
- Carbonyl group
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | Not Available |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Adiposome
- Cell membrane
- Cytoplasm
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0097-9100000000-4296c67313a78a015074 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00y1-9820000000-7256ed8afed29ebf4daa | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0r0r-9200000000-a7d6bf6e90c8c752f69c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0a4j-9000000000-fe9a2716010ffb94cff8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a4l-9000000000-715b545170085466a6b2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0uxr-3900000000-d8f24d02712f429591a7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0zgi-9800000000-0821ba5748054e7de369 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-e5b2f743cb4dd5e99ec5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-5900000000-587bb9b1fdf10856b3a1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05fr-9100000000-a33491d8a46967cdd2d3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-1ec2b5bb3af9f3c925ca | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01b9-8900000000-6e362f577b3d827a8387 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05fr-9100000000-6950ab2628a11bf6839a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9000000000-5278bf25a974c9f30369 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0zgi-9400000000-aee3a280a6df1ae985b6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-45e4e2c8c6ea7216859f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052b-9000000000-845222220a578798d8ce | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|