| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:43:12 UTC |
|---|
| Update Date | 2020-04-22 15:07:20 UTC |
|---|
| BMDB ID | BMDB0001260 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | ADP-Ribosyl-L-arginine |
|---|
| Description | ADP-Ribosyl-L-arginine belongs to the class of organic compounds known as purine nucleotide sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. Based on a literature review very few articles have been published on ADP-Ribosyl-L-arginine. |
|---|
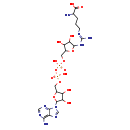
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| N(Omega)-(ADP-D-ribosyl)-arginine | HMDB | | N(Omega)-(ADP-D-ribosyl)-L-arginine | HMDB | | N(Omega)-(ADP-delta-ribosyl)-arginine | HMDB | | N(Omega)-(ADP-delta-ribosyl)-L-arginine | HMDB | | N(W)-(ADP-D-Ribosyl)-arginine | HMDB | | N(W)-(ADP-D-Ribosyl)-L-arginine | HMDB | | N(W)-(ADP-delta-Ribosyl)-arginine | HMDB | | N(W)-(ADP-delta-Ribosyl)-L-arginine | HMDB | | N2-(ADP-D-Ribosyl)-L-arginine | HMDB | | N2-(ADP-delta-Ribosyl)-L-arginine | HMDB | | 2-Amino-5-(n'-{5-[({[({[5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-3,4-dihydroxyoxolan-2-yl}carbamimidamido)pentanoate | Generator, HMDB |
|
|---|
| Chemical Formula | C21H35N9O15P2 |
|---|
| Average Molecular Weight | 715.5014 |
|---|
| Monoisotopic Molecular Weight | 715.172784519 |
|---|
| IUPAC Name | 2-amino-5-[(E)-[amino({5-[({[({[5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-3,4-dihydroxyoxolan-2-yl}amino)methylidene]amino]pentanoic acid |
|---|
| Traditional Name | 2-amino-5-[(E)-{amino[(5-{[({[5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(hydroxy)phosphoryl)oxy]methyl}-3,4-dihydroxyoxolan-2-yl)amino]methylidene}amino]pentanoic acid |
|---|
| CAS Registry Number | 103960-56-1 |
|---|
| SMILES | NC(CCC\N=C(/N)NC1OC(COP(O)(=O)OP(O)(=O)OCC2OC(C(O)C2O)N2C=NC3=C(N)N=CN=C23)C(O)C1O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C21H35N9O15P2/c22-8(20(35)36)2-1-3-25-21(24)29-18-14(33)12(31)9(43-18)4-41-46(37,38)45-47(39,40)42-5-10-13(32)15(34)19(44-10)30-7-28-11-16(23)26-6-27-17(11)30/h6-10,12-15,18-19,31-34H,1-5,22H2,(H,35,36)(H,37,38)(H,39,40)(H2,23,26,27)(H3,24,25,29) |
|---|
| InChI Key | IWVSYNGKNZFSSA-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine nucleotide sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine nucleotide sugars |
|---|
| Direct Parent | Purine nucleotide sugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine nucleotide sugar
- Purine ribonucleoside diphosphate
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- Alpha-amino acid
- Alpha-amino acid or derivatives
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- Monoalkyl phosphate
- Imidolactam
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Alkyl phosphate
- Phosphoric acid ester
- Pyrimidine
- Tetrahydrofuran
- Azole
- Imidazole
- Heteroaromatic compound
- Amino acid or derivatives
- Secondary alcohol
- Amino acid
- Guanidine
- Organoheterocyclic compound
- Carboximidamide
- Carboxylic acid derivative
- Carboxylic acid
- Organic 1,3-dipolar compound
- Monocarboxylic acid or derivatives
- Propargyl-type 1,3-dipolar organic compound
- Azacycle
- Oxacycle
- Hydrocarbon derivative
- Organic oxygen compound
- Primary amine
- Carbonyl group
- Alcohol
- Organonitrogen compound
- Amine
- Organic nitrogen compound
- Primary aliphatic amine
- Organic oxide
- Organopnictogen compound
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00vm-2141819000-b4cd236e4b472d8de1d1 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002r-0910002000-a9b7b3adb57f97d06839 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900000000-d4ae4b923db103b6c0e3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-002r-1900000000-63f0a68905fb8344837e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0089-0910100100-616900a605286795f873 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-1900100000-55d1e9e7a348f9e3bd0f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a59-2900000000-b87c59d767dede055a0b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0400003900-2a6527246116777408bd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dr-5223069200-38cce354551558efeaae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000f-2839100000-e7fd9e6545c90d058b94 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-022i-0000059600-ffcc5f99449300280213 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001l-0000091000-30b0a2fb9a745e4e75da | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a6s-9328634000-e6079b9091ea7d4fb603 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|