| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:55:31 UTC |
|---|
| Update Date | 2020-04-22 15:11:03 UTC |
|---|
| BMDB ID | BMDB0002467 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | A-Ketoglutaric acid oxime |
|---|
| Description | A-Ketoglutaric acid oxime, also known as a-ketoglutarate oxime or 2-oxime-(8ci)-2-oxoglutarate, belongs to the class of organic compounds known as dicarboxylic acids and derivatives. These are organic compounds containing exactly two carboxylic acid groups. Based on a literature review very few articles have been published on A-Ketoglutaric acid oxime. |
|---|
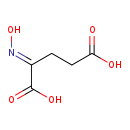
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| a-Ketoglutarate oxime | Generator | | 2-(Hydroxyimino)- pentanedioate | HMDB | | 2-(Hydroxyimino)- pentanedioic acid | HMDB | | 2-Oxime-(8ci)-2-oxoglutarate | HMDB | | 2-Oxime-(8ci)-2-oxoglutaric acid | HMDB | | alpha-Ketoglutaric acid oxime | HMDB | | Oxime-(6ci,7ci)-2-oxoglutarate | HMDB | | Oxime-(6ci,7ci)-2-oxoglutaric acid | HMDB | | (2E)-2-(Hydroxyimino)pentanedioate | HMDB |
|
|---|
| Chemical Formula | C5H7NO5 |
|---|
| Average Molecular Weight | 161.1128 |
|---|
| Monoisotopic Molecular Weight | 161.032422339 |
|---|
| IUPAC Name | (2E)-2-(hydroxyimino)pentanedioic acid |
|---|
| Traditional Name | α-ketoglutaric acid oxime |
|---|
| CAS Registry Number | 2211-15-6 |
|---|
| SMILES | O\N=C(/CCC(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H7NO5/c7-4(8)2-1-3(6-11)5(9)10/h11H,1-2H2,(H,7,8)(H,9,10)/b6-3+ |
|---|
| InChI Key | RSXBEVMDACDZRB-ZZXKWVIFSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dicarboxylic acids and derivatives. These are organic compounds containing exactly two carboxylic acid groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Dicarboxylic acids and derivatives |
|---|
| Direct Parent | Dicarboxylic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty acid
- Dicarboxylic acid or derivatives
- Ketoxime
- Oxime
- Carboxylic acid
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00kf-9700000000-5b68d14aefb4495a7c23 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00y0-9740000000-ec798e8f29f01f1bb847 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-03di-0900000000-cbe03c2f3f7159c84f47 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0229-9300000000-2f48a3637bf6f93b0a4f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-004i-9100000000-0e3c81c755b2c6bbfa17 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-1900000000-a167f1a381e6ad56c282 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-010d-9800000000-5a86d57b066f20c266dd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00b9-9000000000-f6df3f8c6961761cf535 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0900000000-76962cbc4d6f6c76c617 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014v-8900000000-b3e90a09becd81b192a6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-9200000000-70e887624a19f39d7f6b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002f-1900000000-983465a642f7d2572fbc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dj-9300000000-6307fdf0004668afe167 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ab9-9000000000-92f229cf7d2f7ffd21cf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00kb-9600000000-6019fef8199a8149e3f6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-9200000000-66de1dbc6e6d433d84ae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-ab74eca8e41c17224950 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|