| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:04:15 UTC |
|---|
| Update Date | 2020-06-04 20:32:33 UTC |
|---|
| BMDB ID | BMDB0003402 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Pectin |
|---|
| Description | Pectin, also known as methoxy pectin or D-lyxose, belongs to the class of organic compounds known as glucuronic acid derivatives. Glucuronic acid derivatives are compounds containing a glucuronic acid moiety (or a derivative), which consists of a glucose moiety with the C6 carbon oxidized to a carboxylic acid. Pectin exists as a solid, possibly soluble (in water), and an extremely weak basic (essentially neutral) compound (based on its pKa) molecule. Pectin exists in all living species, ranging from bacteria to humans. |
|---|
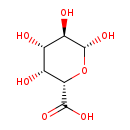
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (+)-Xylose | HMDB | | 2,3,4,5-Tetrahydroxypentanal | HMDB | | D-Lyxose | HMDB | | DL-Xylose | HMDB | | L(+)-Xylose | HMDB | | L-Lyxose | HMDB | | Lyxose | HMDB | | Pectin sugar | HMDB | | Pectinose | HMDB | | Pentose | HMDB | | Trobicin | HMDB | | Pectinic acid | HMDB | | Calcium pectinate | HMDB | | Methoxy pectin | HMDB | | Methoxylpectin | HMDB | | Methoxypectin | HMDB | | Zinc pectinate | HMDB | | Galacturonate | HMDB | | b-D-Galacturonate | HMDB | | b-D-Galacturonic acid | HMDB | | beta-D-Galacturonate | HMDB | | Β-D-galacturonate | HMDB | | Β-D-galacturonic acid | HMDB | | Pectin | MeSH | | b-D-Galactopyranuronate | Generator | | b-D-Galactopyranuronic acid | Generator | | beta-D-Galactopyranuronate | Generator | | Β-D-galactopyranuronate | Generator | | Β-D-galactopyranuronic acid | Generator |
|
|---|
| Chemical Formula | C6H10O7 |
|---|
| Average Molecular Weight | 194.1394 |
|---|
| Monoisotopic Molecular Weight | 194.042652674 |
|---|
| IUPAC Name | (2S,3R,4S,5R,6R)-3,4,5,6-tetrahydroxyoxane-2-carboxylic acid |
|---|
| Traditional Name | β-D-galactopyranuronic acid |
|---|
| CAS Registry Number | 9000-69-5 |
|---|
| SMILES | O[C@@H]1O[C@@H]([C@H](O)[C@H](O)[C@H]1O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H10O7/c7-1-2(8)4(5(10)11)13-6(12)3(1)9/h1-4,6-9,12H,(H,10,11)/t1-,2+,3+,4-,6+/m0/s1 |
|---|
| InChI Key | AEMOLEFTQBMNLQ-DTEWXJGMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glucuronic acid derivatives. Glucuronic acid derivatives are compounds containing a glucuronic acid moiety (or a derivative), which consists of a glucose moiety with the C6 carbon oxidized to a carboxylic acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Glucuronic acid derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glucuronic acid or derivatives
- Beta-hydroxy acid
- Hydroxy acid
- Pyran
- Monosaccharide
- Oxane
- Hemiacetal
- Secondary alcohol
- Carboxylic acid derivative
- Carboxylic acid
- Oxacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Polyol
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected and Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fvi-3900000000-623cfb10797e088f31a3 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (5 TMS) - 70eV, Positive | splash10-000l-6242950000-7b610ed98440b47c2a32 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-00kr-6900000000-a4a0a279e2e16f432677 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-000i-9100000000-1c90dbe22f830f93ad0f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-004i-9000000000-cefd20d23fb5136d2104 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002b-0900000000-142e51c19f7b152d18e6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-056s-1900000000-80978d6e9cf43c2a99ae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4r-9400000000-fe9d58fcfd8f4880243e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0007-2900000000-eecfa94a9428872c9b56 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000w-4900000000-c10f02b5bf83d62924f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9200000000-21b195d29543b18b8595 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0900000000-2877a7e6d3ecd32cf172 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000b-7900000000-d748e59637b495442c9e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-9100000000-40ac961f184de816ed86 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-3900000000-64f3d0eb26f7bc8ca624 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a73-9300000000-329488f791f643b1c308 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-474e82a5911096bc3b39 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| General References | - Klein MS, Almstetter MF, Schlamberger G, Nurnberger N, Dettmer K, Oefner PJ, Meyer HH, Wiedemann S, Gronwald W: Nuclear magnetic resonance and mass spectrometry-based milk metabolomics in dairy cows during early and late lactation. J Dairy Sci. 2010 Apr;93(4):1539-50. doi: 10.3168/jds.2009-2563. [PubMed:20338431 ]
- Sun HZ, Shi K, Wu XH, Xue MY, Wei ZH, Liu JX, Liu HY: Lactation-related metabolic mechanism investigated based on mammary gland metabolomics and 4 biofluids' metabolomics relationships in dairy cows. BMC Genomics. 2017 Dec 2;18(1):936. doi: 10.1186/s12864-017-4314-1. [PubMed:29197344 ]
|
|---|