| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:20:01 UTC |
|---|
| Update Date | 2020-06-04 20:22:56 UTC |
|---|
| BMDB ID | BMDB0006050 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | o-Tyrosine |
|---|
| Description | O-Tyrosine, also known as DL- O-tyrosine or ortho-tyrosine, belongs to the class of organic compounds known as phenylalanine and derivatives. Phenylalanine and derivatives are compounds containing phenylalanine or a derivative thereof resulting from reaction of phenylalanine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. O-Tyrosine exists as a solid, possibly soluble (in water), and a very strong basic compound (based on its pKa) molecule. |
|---|
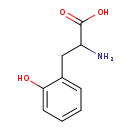
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Amino-3-(2-hydroxyphenyl)propanoic acid | ChEBI | | 2-Amino-3-(2-hydroxyphenyl)propanoate | Generator | | 2-Hydroxy-DL-phenylalanine | HMDB | | 2-Hydroxy-phenylalanine | HMDB | | 2-Hydroxyphenylalanine | HMDB | | 2-Tyrosine | HMDB | | 3-(O-Hydroxyphenyl)DL-alanine | HMDB | | DL- O-Tyrosine | HMDB | | DL-3-(O-Hydroxyphenyl)alanine | HMDB | | DL-O-Tyrosine | HMDB | | Ortho-tyrosine | HMDB | | 2-Tyrosine, (D)-isomer | HMDB | | 2-Tyrosine, (DL)-isomer | HMDB | | 2-Tyrosine, (L)-isomer | HMDB |
|
|---|
| Chemical Formula | C9H11NO3 |
|---|
| Average Molecular Weight | 181.1885 |
|---|
| Monoisotopic Molecular Weight | 181.073893223 |
|---|
| IUPAC Name | 2-amino-3-(2-hydroxyphenyl)propanoic acid |
|---|
| Traditional Name | 2-tyrosine |
|---|
| CAS Registry Number | 2370-61-8 |
|---|
| SMILES | NC(CC1=CC=CC=C1O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C9H11NO3/c10-7(9(12)13)5-6-3-1-2-4-8(6)11/h1-4,7,11H,5,10H2,(H,12,13) |
|---|
| InChI Key | WRFPVMFCRNYQNR-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phenylalanine and derivatives. Phenylalanine and derivatives are compounds containing phenylalanine or a derivative thereof resulting from reaction of phenylalanine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Phenylalanine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phenylalanine or derivatives
- 3-phenylpropanoic-acid
- Amphetamine or derivatives
- Alpha-amino acid
- 1-hydroxy-2-unsubstituted benzenoid
- 1-hydroxy-4-unsubstituted benzenoid
- Aralkylamine
- Phenol
- Monocyclic benzene moiety
- Benzenoid
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxygen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Hydrocarbon derivative
- Primary aliphatic amine
- Organic nitrogen compound
- Carbonyl group
- Organic oxide
- Organopnictogen compound
- Amine
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected and Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 262 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 2 mg/mL at 17 °C | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-4900000000-aa81f75d9ed4e977df68 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0a4i-5491000000-9a9969815e032107fc44 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-001i-0900000000-ebb473b1eb9270c27a26 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-015i-0900000000-d3b4c59468db65e33b80 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00kf-9700000000-cf7cdfd040ca5a451fa2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0019-0900000000-01ee9c831970c7f79ede | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900000000-04fdaa25b741b52c551e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056r-9300000000-639776ec42cd7c70b706 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-1900000000-9911c42139ecbba3b834 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05fr-7900000000-b4167e456277ba9ec5ba | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fu-9200000000-159e5a860c481d106109 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00lr-0900000000-0789c3bec09b7b71842e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0aor-2900000000-294b1d1404168e5de571 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05mo-9800000000-9ac4c7e756ac9d42572d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00li-0900000000-1d0af1b840230361c6f8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0aou-2900000000-66a3f07b95f44b6d0341 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9100000000-06c2ec0981d6e289ea4f | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| |
| Blood | Detected and Quantified | 91 +/- 10 uM | Not Specified | Not Specified | Normal | | details | | Liver | Detected and Quantified | 84 +/- 27 nmol/g of tissue | Not Specified | Not Specified | Normal | | details | | Longissimus Thoracis Muscle | Detected and Quantified | 54 +/- 14 nmol/g of tissue | Not Specified | Not Specified | Normal | | details | | Milk | Detected but not Quantified | Not Quantified | Not Specified | Not Specified | Normal | | details | | Ruminal Fluid | Detected and Quantified | 295 +/- 136 uM | Not Specified | Not Specified | Normal | | details | | Semimembranosus Muscle | Detected and Quantified | 52 +/- 10 nmol/g of tissue | Not Specified | Not Specified | Normal | | details | | Testis | Detected and Quantified | 43 +/- 9 nmol/g of tissue | Not Specified | Not Specified | Normal | | details |
|
|---|