| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 18:22:58 UTC |
|---|
| Update Date | 2020-04-22 15:45:59 UTC |
|---|
| BMDB ID | BMDB0011690 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 7-Aminomethyl-7-carbaguanine |
|---|
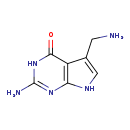
| Description | 7-Aminomethyl-7-carbaguanine, also known as 7-deaza-7-aminomethyl-guanine or PREQ1, belongs to the class of organic compounds known as pyrrolo[2,3-d]pyrimidines. These are aromatic heteropolycyclic compounds containing a pyrrolo[2,3-d]pyrimidine ring system, which is an pyrrolopyrimidine isomers having the 3 ring nitrogen atoms at the 1-, 5-, and 7-positions. 7-Aminomethyl-7-carbaguanine is a very strong basic compound (based on its pKa). 7-Aminomethyl-7-carbaguanine exists in all living organisms, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 7-DEAZA-7-aminomethyl-guanine | ChEBI | | 7-Aminomethyl-7-deazaguanine | Kegg | | PreQ1 | HMDB |
|
|---|
| Chemical Formula | C7H9N5O |
|---|
| Average Molecular Weight | 179.1793 |
|---|
| Monoisotopic Molecular Weight | 179.080709935 |
|---|
| IUPAC Name | 2-amino-5-(aminomethyl)-3H,4H,7H-pyrrolo[2,3-d]pyrimidin-4-one |
|---|
| Traditional Name | 7-aminomethyl-7-deazaguanine |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | NCC1=CNC2=C1C(=O)NC(N)=N2 |
|---|
| InChI Identifier | InChI=1S/C7H9N5O/c8-1-3-2-10-5-4(3)6(13)12-7(9)11-5/h2H,1,8H2,(H4,9,10,11,12,13) |
|---|
| InChI Key | MEYMBLGOKYDGLZ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrrolo[2,3-d]pyrimidines. These are aromatic heteropolycyclic compounds containing a pyrrolo[2,3-d]pyrimidine ring system, which is an pyrrolopyrimidine isomers having the 3 ring nitrogen atoms at the 1-, 5-, and 7-positions. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pyrrolopyrimidines |
|---|
| Sub Class | Pyrrolo[2,3-d]pyrimidines |
|---|
| Direct Parent | Pyrrolo[2,3-d]pyrimidines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrrolo[2,3-d]pyrimidine
- Aralkylamine
- Hydroxypyrimidine
- Substituted pyrrole
- Pyrimidine
- Heteroaromatic compound
- Pyrrole
- Azacycle
- Organic nitrogen compound
- Hydrocarbon derivative
- Amine
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Organopnictogen compound
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-06vr-1900000000-6c74d2d331883324fbd8 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03e9-0900000000-186cf56f77e8ac46162e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0900000000-e42a8c4daace1afde7ae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0229-1900000000-d9d17cbc8d643eb3bb92 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0900000000-7c00db685d48eeeaa166 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-2900000000-8a8ba88dc9cf497add5e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9600000000-8d2f3dc2af852732e95b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0900000000-640c17980dbf68c2f745 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03dj-1900000000-ed2d82c910ac3948e2e7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9700000000-110463c1e8dbf082afcf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0900000000-056d9eb91af27afcf50d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0900000000-8b141eab907933d865b1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0075-4900000000-b0875f15fc961a04cee6 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|