| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2018-07-17 17:24:24 UTC |
|---|
| Update Date | 2020-04-22 15:51:48 UTC |
|---|
| BMDB ID | BMDB0062269 |
|---|
| Secondary Accession Numbers | - BMDB0064145
- BMDB62269
- BMDB64145
|
|---|
| Metabolite Identification |
|---|
| Common Name | Valylproline |
|---|
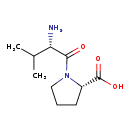
| Description | Valylproline, also known as V-P or L-val-L-pro, belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. Based on a literature review a small amount of articles have been published on Valylproline. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| L-Val-L-pro | ChEBI | | V-P | ChEBI | | VP | ChEBI | | 1-L-Valyl-L-proline | HMDB | | 1-L-Valylproline | HMDB | | L-Valyl-L-proline | HMDB | | L-alpha-Valyl-L-proline | HMDB | | L-Α-valyl-L-proline | HMDB | | N-L-Valyl-L-proline | HMDB | | N-Valylproline | HMDB | | V-p Dipeptide | HMDB | | VP Dipeptide | HMDB | | Val-pro | HMDB | | Valine proline dipeptide | HMDB | | Valine-proline dipeptide | HMDB | | Valyl-proline | HMDB | | Valylproline | ChEBI |
|
|---|
| Chemical Formula | C10H18N2O3 |
|---|
| Average Molecular Weight | 214.265 |
|---|
| Monoisotopic Molecular Weight | 214.131742448 |
|---|
| IUPAC Name | (2S)-1-[(2S)-2-amino-3-methylbutanoyl]pyrrolidine-2-carboxylic acid |
|---|
| Traditional Name | (2S)-1-[(2S)-2-amino-3-methylbutanoyl]pyrrolidine-2-carboxylic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CC(C)[C@H](N)C(=O)N1CCC[C@H]1C(O)=O |
|---|
| InChI Identifier | InChI=1S/C10H18N2O3/c1-6(2)8(11)9(13)12-5-3-4-7(12)10(14)15/h6-8H,3-5,11H2,1-2H3,(H,14,15)/t7-,8-/m0/s1 |
|---|
| InChI Key | GIAZPLMMQOERPN-YUMQZZPRSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Dipeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-dipeptide
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Valine or derivatives
- Proline or derivatives
- N-acyl-l-alpha-amino acid
- Alpha-amino acid amide
- Alpha-amino acid or derivatives
- N-acylpyrrolidine
- Pyrrolidine carboxylic acid
- Pyrrolidine carboxylic acid or derivatives
- Pyrrolidine
- Tertiary carboxylic acid amide
- Amino acid or derivatives
- Carboxamide group
- Amino acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Azacycle
- Organoheterocyclic compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Organic oxide
- Organopnictogen compound
- Primary aliphatic amine
- Organic nitrogen compound
- Carbonyl group
- Amine
- Hydrocarbon derivative
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | -2.22 | Extrapolated |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0190000000-a2b677fc4252b284c757 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-4900000000-fbea73dc727349bb2d7c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03dj-9500000000-45b2fc983242246e181b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-2980000000-2a6ecccc6a1cb4556002 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01b9-9510000000-8b1f1a971d05a351af08 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9000000000-7efa18764ade27937c96 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|